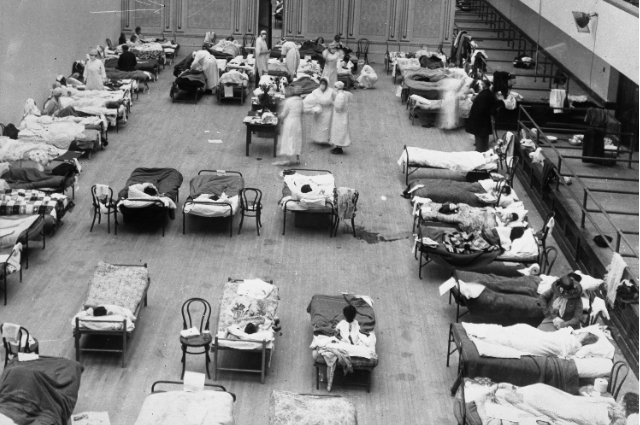
In the annals of human history, few events have left as profound a mark as the 1918 influenza pandemic, often referred to as the "Spanish Flu." This devastating outbreak, which swept across the globe from 1918 to 1920, claimed an estimated 20 to 100 million lives—surpassing the death toll of World War I. Its severity and mysterious origins have long captivated scientists and historians, driving efforts to understand what made this virus so deadly.
In July 2025, a groundbreaking discovery brought us closer to answering that question. Researchers from the Universities of Basel and Zurich successfully decoded the genome of the 1918 flu virus using a century-old tissue sample preserved in the University of Zurich’s Medical Collection. This remarkable achievement, led by paleogeneticist Verena Schünemann, offers unprecedented insights into one of history’s deadliest pandemics and holds valuable lessons for preparing for future global health crises. This article explores the discovery, its findings, and its implications for science and society.
The 1918 Pandemic: A Historical Catastrophe
The 1918 influenza pandemic was unlike any other. While most flu outbreaks primarily affect the very young and elderly, the Spanish Flu disproportionately struck healthy young adults, a phenomenon that puzzled scientists for decades. The pandemic unfolded in three waves: a milder first wave in spring 1918, a devastating second wave in fall 1918, and a third wave in spring 1919. The second wave was particularly lethal, with mortality rates soaring as entire communities were overwhelmed.
The social and economic impacts were staggering. Public gatherings were banned, schools and businesses shuttered, and mask-wearing became mandatory in many regions—measures that echo responses to modern pandemics like COVID-19. In Switzerland alone, the pandemic infected at least 2 million people and caused approximately 25,000 deaths, with young men particularly affected during the second wave. The psychological toll was equally profound, leaving a lasting imprint on survivors and shaping public health policies for generations.
The Discovery: A Window into the Past
The journey to decode the 1918 flu virus genome began with a formalin-fixed wet specimen from the University of Zurich’s Medical Collection. This sample, taken from an 18-year-old patient who died in Zurich during the first wave in July 1918, had been preserved for over a century, offering a rare opportunity to study the virus’s genetic makeup.
The research was led by Verena Schünemann, a paleogeneticist and professor of archaeological science at the University of Basel, who previously worked at the University of Zurich. Her team, including first author Christian Urban from UZH, faced the daunting challenge of extracting and sequencing ancient RNA fragments, which are notoriously unstable and prone to degradation. To overcome this, Urban developed an innovative ligation-based RNA workflow that enhanced the recovery of small RNA fragments, a significant advancement in paleogenomics.
“This is the first time we’ve had access to an influenza genome from the 1918–1920 pandemic in Switzerland. It opens up new insights into the dynamics of how the virus adapted in Europe at the start of the pandemic,” Schünemann said, highlighting the study’s significance University of Zurich News.
The team successfully reconstructed the first Swiss genome of the 1918 influenza virus, achieving high coverage (63.9× average, with 98.8% of the genome covered at least once). This genome revealed that the virus had already developed key adaptations to infect humans early in the pandemic, shedding light on its rapid spread and lethality.
Key Findings: The Virus’s Lethal Adaptations
The decoded genome provided critical insights into the virus’s ability to infect humans. The researchers identified three key mutations that likely contributed to its virulence:
Mutation Gene Effect
| Mutation | Gene | Effect |
D16, P283 | Nucleoprotein (NP) | Increased resistance to the human immune system’s antiviral component (MxA) |
| D222 | Hemagglutinin (HA) | Enhanced binding to human cell receptors, improving infectivity |
These mutations were present in the Swiss strain from the outset and persisted throughout the pandemic, indicating early and effective adaptation to human hosts. The study also compared the Swiss genome to previously sequenced genomes from Germany and North America, revealing that the 1918 virus was part of a diverse viral population with multiple genotypes circulating simultaneously. The Swiss strain differed from a Munich sample (MU-162) at 35 positions, with 14 resulting in amino acid changes, though its HA segment was identical to MU-162.
Phylogenetic analysis showed that the Swiss genome clustered with strains from mainland Europe and North America from both the first and second waves. Notably, the 1918 influenza A virus (IAV) exhibited greater genetic diversity than the 2009 H1N1 virus in certain genome segments (PB2, PA, and HA), suggesting a complex evolutionary history.
The Science Behind the Breakthrough
The study’s success hinged on a novel method developed by Christian Urban, which improved the recovery of ancient RNA fragments from formalin-fixed specimens. This ligation-based workflow outperformed previous techniques, offering a new tool for studying historical pathogens. The data from this study is publicly available under BioProject ID: PRJNA1181848, enabling further research by the global scientific community.
Collaborating institutions, including the University of Zurich’s Medical Collection and the Berlin Museum of Medical History at Charité University Hospital, played a crucial role in providing access to historical samples. The study, published on July 1, 2025, in BMC Biology under the title “An ancient influenza genome from Switzerland allows deeper insights into host adaptation during the 1918 flu pandemic in Europe,” marks a significant milestone in paleogenomics BMC Biology.
Historical and Scientific Significance
This discovery is a landmark in understanding the 1918 pandemic. By decoding the virus’s genome, researchers have gained insights into its pathogenicity and transmissibility, key factors in its devastating impact. The study confirms that the virus likely originated from an avian strain
that adapted to humans, a finding consistent with earlier research Nature, 2005. The identification of specific mutations provides clues about why the virus was so lethal, particularly its ability to trigger a cytokine storm—an overactive immune response that caused severe lung damage.
The research also highlights the value of preserved medical collections. These archives, often overlooked, serve as time capsules that allow scientists to study pathogens from the past. By comparing the 1918 virus to modern strains, researchers can better understand viral evolution and develop models to predict future pandemic risks.
Implications for Future Pandemic Preparedness
The lessons from the 1918 pandemic remain highly relevant today. Understanding how viruses adapt to humans is critical for developing effective countermeasures, such as vaccines and antiviral therapies. The ability to sequence ancient viral genomes opens new avenues for research, enabling scientists to track pathogen evolution over time and anticipate how emerging viruses might behave.
This study’s findings could inform strategies for monitoring and combating future pandemics. For example, identifying mutations that enhance infectivity or immune evasion can guide the development of targeted interventions. The research also underscores the importance of international collaboration and data sharing, as seen in the public availability of the study’s data.
Comparing the 1918 Pandemic to Modern Outbreaks
The 1918 flu shares similarities with modern pandemics, such as the 2009 H1N1 swine flu and COVID-19, but its extreme virulence sets it apart. Unlike typical flu viruses, the 1918 strain caused severe pneumonia and high mortality among young adults, likely due to its ability to induce a cytokine storm. By contrast, the 2009 H1N1 virus was less deadly, and COVID-19 has affected a broader age range. Studying the 1918 virus provides a benchmark for understanding the factors that drive pandemic severity.
| Pandemic | Years | Estimated Deaths | Key Characteristics |
1918 Spanish Flu | 1918–1920 | 20–100 million | High mortality in young adults, rapid global spread |
| 2009 H1N1 | 2009–2010 | 151,700–575,400 | Milder, affected all age groups |
| COVID-19 | 2019–present | 6.9 million (as of 2023) | Broad age impact, significant social disruption |
Conclusion: Learning from the Past to Protect the Future
The decoding of the 1918 flu virus genome is a testament to the power of scientific innovation and the enduring value of historical preservation. By unraveling the genetic secrets of this deadly virus, researchers have deepened our understanding of one of history’s greatest tragedies and provided a foundation for preparing for future pandemics. As we navigate an era of emerging infectious diseases, the lessons from 1918 remind us of the importance of vigilance, research, and global cooperation.
In the words of Verena Schünemann, this discovery “opens up new insights into the dynamics of how the virus adapted in Europe at the start of the pandemic.” As science continues to advance, studies like this will play a crucial role in safeguarding public health and ensuring that the horrors of 1918 are not repeated.